Wednesday, May 21, 2008 - 9:45 AM
Medical Arts Building, Rm M-136 (Queensborough Community College)
586
Conformational Analysis of cADPR and cADPR Analog Agonists and Antagonists Using PSEUROT, Molecular Mechanics, and Ab Inito Calculations
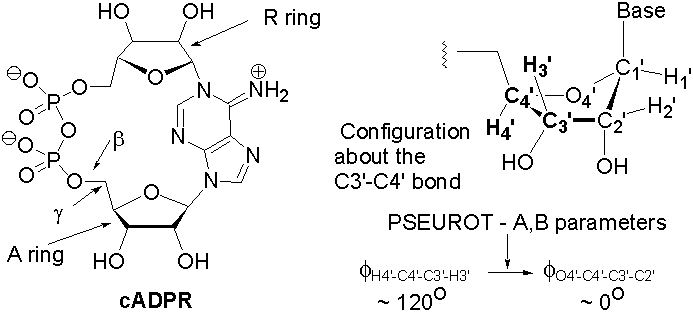
Cyclic adenosine diphosphate ribose (cADPR), a cyclic metabolite of NAD+, is a second messenger that causes release of calcium from intracellular stores. For some time our lab has been attempting to unravel the structure-activity relationships in cADPR and cADPR analogs, in particular the conformation of the 5-membered furanose rings. The key feature of the PSEUROT program is its ability to convert 1H-1H NMR coupling constants, via a Karplus equation, to exocyclic H-C-C-H torsion angles (φexo); these angles are then converted to the endocyclic ring torsions (φendo) needed to describe the conformation of the furanose (φexo = A•φendo + B). The �A� and �B� parameters are specific to a particular furanose configuration (e.g. ribose, arabinose, deoxyribose, etc.) and are supplied by the PSEUROT program, at least for the common furanoses. In the event that a particular furanose is not in the PSEUROT database, or if one suspects the supplied parameters may not be appropriate (as in cADPR), then the �A� and �B� parameters must be calculated, typically from a survey of in silico structures obtained from molecular modeling. This talk will focus on our efforts to determine the appropriate �A� and �B� parameters for cADPR via molecular mechanics, semi-empirical, and ab initio calculations, and their effect on the PSEUROT calculations.